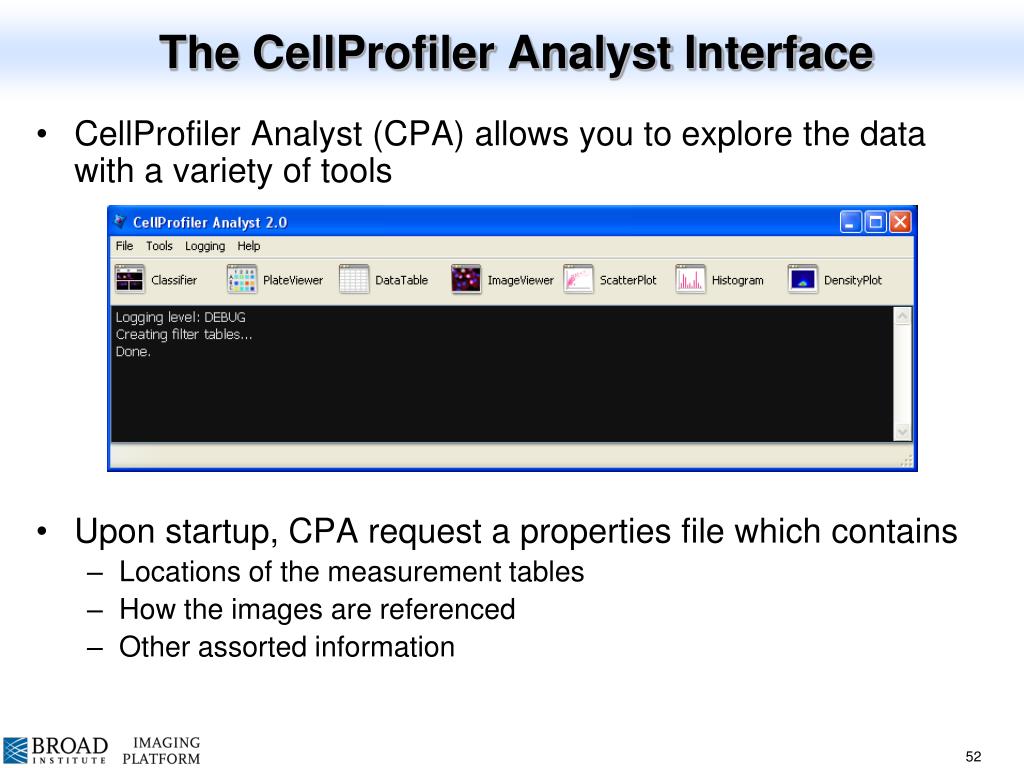
This file was exported by CellProfiler’s ExportToDatabase module. Run the CPAnalyst file created by the install process above. A dialog will appear asking you to select a properties file. Navigate to the cpaexample directory and select the example.properties file. You’re now ready to experiment with CellProfiler Analyst! CellProfiler Analyst now includes several machine learning algorithm options; for our purposes, we chose a GradientBoosting classifier and a Random Forest classifier. We picked boosting in order to compare with the boosting results in (Blasi et al., 2016), and Random Forests as a second approach often considered best-in-class.
exports data directly to a database, or in database readable format, including an imported file with column names and a CellProfiler Analyst properties file, if desired. This module exports measurements directly to a database or to a SQL-compatible format. It allows you to create and import MySQL and associated data files into a database and gives you the option of creating a properties file for use with CellProfiler Analyst. Optionally, you can create an SQLite database file if you do not have a server on which to run MySQL itself.This module must be run at the end of a pipeline, or second to last if you are using the CreateBatchFiles module. If you forget this module, you can also run the ExportDatabase data tool after processing is complete; its functionality is the same.
The database is set up with two primary tables. Parsnips and carrots recipes. These tables are the Per_Image table and the Per_Object table (which may have a prefix if you specify):
Cellprofiler Analyst Classifier
- The Per_Image table consists of all the per-image measurements made during the pipeline, plus per-image population statistics (such as mean, median, and standard deviation) of the object measurements. There is one per_image row for every 'cycle' that CellProfiler processes (a cycle is usually a single field of view, and a single cycle usually contains several image files, each representing a different channel of the same field of view).
- The Per_Object table contains all the measurements for individual objects. There is one row of object measurements per object identified. The two tables are connected with the primary key column ImageNumber, which indicates the image to which each object belongs. The Per_Object table has another primary key called ObjectNumber, which is unique to each image.
Typically, if multiple types of objects are identified and measured in a pipeline, the numbers of those objects are equal to each other. For example, in most pipelines, each nucleus has exactly one cytoplasm, so the first row of the Per-Object table contains all of the information about object #1, including both nucleus- and cytoplasm-related measurements. If this one-to-one correspondence is not the case for all objects in the pipeline (for example, if dozens of speckles are identified and measured for each nucleus), then you must configure ExportToDatabase to export only objects that maintain the one-to-one correspondence (for example, export only Nucleus and Cytoplasm, but omit Speckles).
If you have extracted 'Plate' and 'Well' metadata from image filenames or loaded 'Plate' and 'Well' metadata via the Metadata or LoadData modules, you can ask CellProfiler to create a 'Per_Well' table, which aggregates object measurements across wells. This option will output a SQL file (regardless of whether you choose to write directly to the database) that can be used to create the Per_Well table. At the secure shell where you normally log in to MySQL, type the following, replacing the italics with references to your database and files:
mysql -h hostname -u username -p databasename < pathtohttp://d1zymp9ayga15t.cloudfront.net/images/perwellsetupfile.SQL

The commands written by CellProfiler to create the Per_Well table will be executed.
Oracle is not fully supported at present; you can create your own Oracle DB using the .csv output option and writing a simple script to upload to the database.
Available measurements
For details on the nomenclature used by CellProfiler for the exported measurements, see Help > General Help > How Measurements Are Named.See also ExportToSpreadsheet.
CellProfiler
CellProfiler is a free open-source software designed to enable biologists without training in computer vision or programming to quantitatively measure phenotypes from thousands of images automatically. More information can be found in the CellProfiler Wiki.
Download CellProfiler.
CellProfiler Analyst
CellProfiler Analyst allows interactive exploration and analysis of data, particularly from high-throughput, image-based experiments. Included is a supervised machine learning system which can be trained to recognize complex and subtle phenotypes, for automatic scoring of millions of cells.
Cell Counting Software

Download CellProfiler Analyst.
DeepProfiler
DeepProfiler is a set of tools that allow you to use deep learning for analyzing imaging data in high-throughput biological experiments. Please, see our Wiki documentation page for more details about how to use it.
Instructions to download DeepProfiler can be found on the Github page.
Piximi
A web-based deep learning tool for classification of human cells, created with Tensorflow.js and React.
